nm4bl2111 – MICROFLUIDIC DEVICE FOR THE DYNAMIC CHARACTERIZATION OF CELL ADHESION USING SINGLE CELL ADHESION DOT ARRAYS (SCADA)
NM4BL-ACS-2021
MICROFLUIDIC DEVICE FOR THE DYNAMIC CHARACTERIZATION OF CELL ADHESION USING SINGLE CELL ADHESION DOT ARRAYS (SCADA)
ABSTRACT
Cell affinity to certain substrates is of great importance for a wide range of applications. One way to quantify it is to analyze single cell arrays as digital indicators of adhesion to the substrate by the presence or absence of cell on a dot. So far, this methodology –Single Cell Adhesion Dot Array or SCADA– has only been exploited on conventional culture plates, whose use restricts time resolution and requires large amounts of cells and reagents. By integrating SCADA substrates into a microfluidic device, we have achieved the rapid, real-time and automatized monitoring of cell adhesion kinetics.
KEYWORDS: cell-based biosensors, single cell, cell adhesion, microfluidic integration.
INTRODUCTION
Cell-substrate interactions are of utmost interest in molecular and cellular biology as well as in the development of biocompatible substrates. The use of single cell arrays to evaluate the degree of affinity between a certain cell type and a given substance provides a simple yet high-throughput tool. A method to isolate cells is the generation of cell adhesion spots with single cell size, which is the basis of the SCADA technology. SCADA assays have proven useful for the characterization of integrin profiles and cell-biomaterial interactions1; although they have also been used for cytotoxicity assays2, confirming their versatility. This method has previously been exploited on culture plates. However, the handling and analysis of culture plates is time-consuming and leads to low time accuracy, therefore, it could greatly benefit from automation. In this work, we present the integration of SCADA substrates within microfluidics to enable dynamic cell adhesion assays.
EXPERIMENTAL
The microfluidic chamber was designed to render a homogeneous velocity of the cell suspension and promote adhesion to the patterned substrate (Figure 1A). The bottom layer was a glass slide, the chamber was defined by white PSA and the top layer was PMMA (Figure 1B). Glass was chosen as the substrate to be patterned because it is transparent and provides the optimal optic requirements for micropatterning by Alvéole’s PRIMO® micropatterning device. The pattern consisted of 20 μm diameter dots of fibronectin (as cell adhesion molecule) and fluorescent BSA-TAMRA (in order to visualize the formation of the pattern) (Figure 1C). Human hair follicle derived Mesenchymal Stromal Cells (hf-MSCs) at a concentration of 2 105 cells mL-1 were introduced into the microfluidic chip at a flow rate of 50 μL min-1. Adhesion was monitored in real-time through direct microscopic observation (Figure 1D) and it was quantified as the percentage of potential adhesion dots that had been occupied by a cell: Dot Array Occupancy or DAO (Figure 1E).
RESULTS AND DISCUSSION
Previous SCADA assays required up to 1 h of cell incubation over the pattern to achieve a DAO above 80 %. In contrast, with the substrates created by the PRIMO® system and flow incubation, a DAO close to 90 % was obtained after only 10 min (Figure 1E). It must be noted that the surface chemistry employed for the formation of
the adhesion islets by this system is different from the one previously developed for the culture plates. In this case, the contrast achieved between the adhesion and the blocked spaces on the surface is much higher, greatly increasing cell affinity/repulsion strengths on the desired spots. This, added to the improved interaction of the cell suspension with the patterned substrate obtained by applying a microfluidic flow, results in the enhanced adhesion kinetic we report. We have proven that translating SCADA assays into a microfluidic device (SCADA-on-chip) not only reduces the usage of materials, reagents and cells, but it also decreases the assay time by a 6-fold. Moreover, the dynamic monitoring of a single chamber in real-time drastically improves time resolution (which is only limited by the time resolution of the camera), directly enhancing the precision of our analysis.
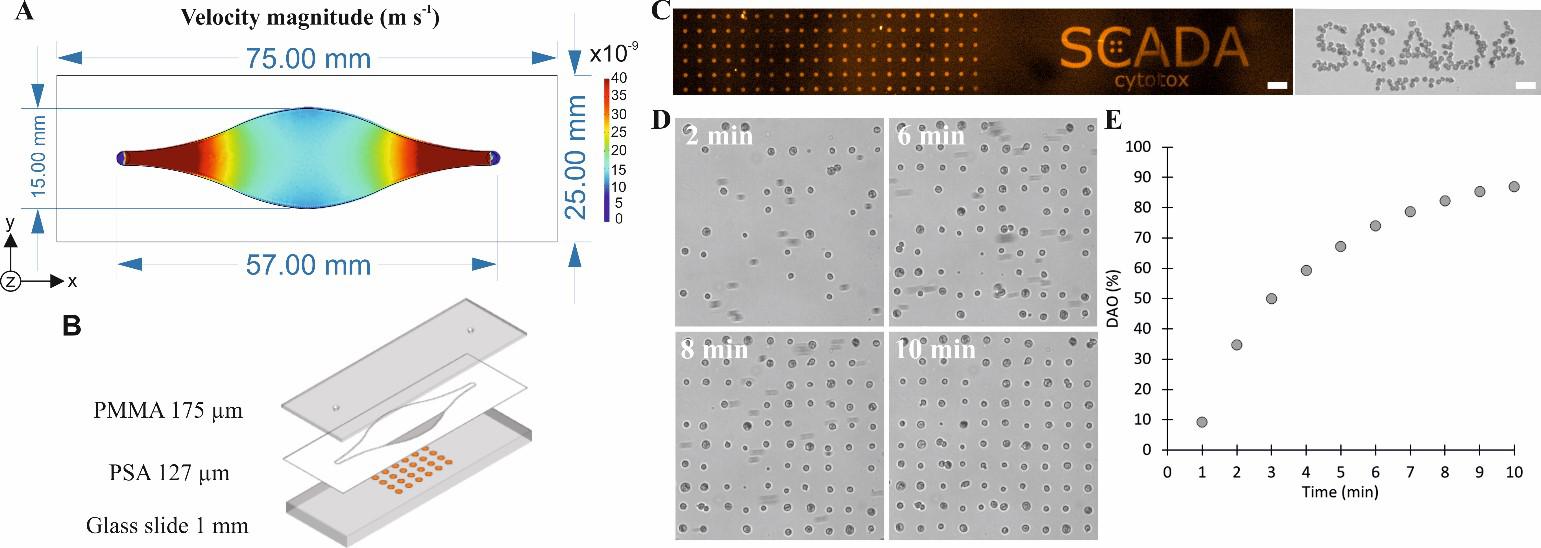
Figure 1. SCADA-on-chip description and results. A) Dimensions of the inner chamber of the chip and COMSOL multiphysics® simulation of the velocities inside the chip when applying a constant flow rate of 50 μL min-1. B) Layer-by-layer representation of the microfluidic chip; the bottom layer was patterned with dots of fibronectin and BSA-TAMRA of 20 μm in diameter. C) Fluorescence image of the dot array and SCADA logo, and brightfield image of cells adhered to the SCADA logo. D) Brightfield images of the evolution of the single cell array after 2, 6, 8 and 10 min of flowing cells over the patterned substrate. E) Adhesion kinetics of hf-MSCs in the SCADA-on-chip device. Scale bars: 100 μm.
CONCLUSION
Considering all this, we can conclude that the developed SCADA-on-chip provides a useful tool for cell adhesion studies. This platform could be applied to any cell type and molecular interaction of interest, where the amount of adhered single cells would be easily and precisely quantified in a short assay time.
ACKNOWLEDGEMENTS
Authors acknowledge funding support from Basque Government, under Grupos Consolidados with Grant No. IT1271-19 and from Gobierno de España, MINECO, with Grant No. BIO2016-80417-P (AEI/FEDER, UE). Authors thank Dr. Marian M. de Pancorbo from BIOMICS group and Dr. Alberto Gorrochategui from “Clínica Dermatológica Ercilla (Bilbao)” for providing the hair follicles from where MSCs were extracted. Authors thank ©Alvéole and distributor Izasa Scientific S. L. U. for the use of the PRIMO® system.
REFERENCES
[1] Gonzalez-Pujana et al., “Extracellular Matrix protein microarray-based biosensor with single cell resolution: Integrin profiling and characterization of cell-biomaterial interactions,” Sensors and Actuators B: Chemical, 299, 126954, 2020.
[2] Garcia-Hernando et al., “Optical single cell resolution cytotoxicity biosensor based on single cell adhesion dot arrays,” Analytical Chemistry, 92, 9658-9665, 2020.